 Parental genomic imprinting is an epigenetic mechanism of gene regulation that results in mono-allelic, parent-of-origin-dependent expression of about 150 genes in eutherian mammals and a handful of genes in metatherians (Barlow & Bartolomei, Cold Spring Harb Perspect Biol, 2014; Ferguson-Smith, Nat Rev Genet, 2011). Genomic imprinting has not been demonstrated in egg-laying mammals (prototherians) and other clades (birds, reptiles, amphibians, fishes). The rationale for the selection of this atypical mechanism of gene regulation during mammalian evolution is still debated (Patten et al., Heredity (Edinb), 2014).
Parental genomic imprinting is an epigenetic mechanism of gene regulation that results in mono-allelic, parent-of-origin-dependent expression of about 150 genes in eutherian mammals and a handful of genes in metatherians (Barlow & Bartolomei, Cold Spring Harb Perspect Biol, 2014; Ferguson-Smith, Nat Rev Genet, 2011). Genomic imprinting has not been demonstrated in egg-laying mammals (prototherians) and other clades (birds, reptiles, amphibians, fishes). The rationale for the selection of this atypical mechanism of gene regulation during mammalian evolution is still debated (Patten et al., Heredity (Edinb), 2014).
The mechanisms that account for the mono-allelic, parent-of-origin-dependent expression of imprinted genes have been studied in great details (Hanna & Kelsey, Heredity (Edinb), 2014; Ferguson-Smith, Nat Rev Genet, 2011). A large number of imprinted genes are clustered at a limited number of imprinted loci. The repression of one of their parental alleles is controlled by the so-called Imprinting Control Regions (ICRs), which usually comprise a Differentially Methylated Region (DMR). The (de)methylation of the paternal and maternal DMRs governs the expression/repression of the neighboring imprinted genes.
 Our team focus on the functions of imprinted genes and aims at identifying the biological processes they control. This is mostly motivated by the numerous human syndroms that result from imprinting defects or cytogenetic alterations of imprinted loci (Mackay & Temple, Eur J Med Genet, 2017). The phenotypes of these syndroms are complex and diverse (Peters, Nat Rev Genet, 2014); this suggests that imprinted genes have diverse functions and may not be functionally related. On the other hand, imprinted gene mutants display common features, i.e. embryonic growth defects and dysregulated energy balance (Plasschaert & Bartolomei, Development, 2014).
Our team focus on the functions of imprinted genes and aims at identifying the biological processes they control. This is mostly motivated by the numerous human syndroms that result from imprinting defects or cytogenetic alterations of imprinted loci (Mackay & Temple, Eur J Med Genet, 2017). The phenotypes of these syndroms are complex and diverse (Peters, Nat Rev Genet, 2014); this suggests that imprinted genes have diverse functions and may not be functionally related. On the other hand, imprinted gene mutants display common features, i.e. embryonic growth defects and dysregulated energy balance (Plasschaert & Bartolomei, Development, 2014).
One possible interpretation is that imprinted genes are fulfilling different functions in different cell types as depicted in figure 1. In this view, each imprinted gene is proposed to have a key function in the tissue(s) most affected by the corresponding mutant. The control of organismal growth would be accomplished by a whole set of seemingly diverse processes, as suggested by the lack of common Gene Ontology terms associated with imprinted genes.
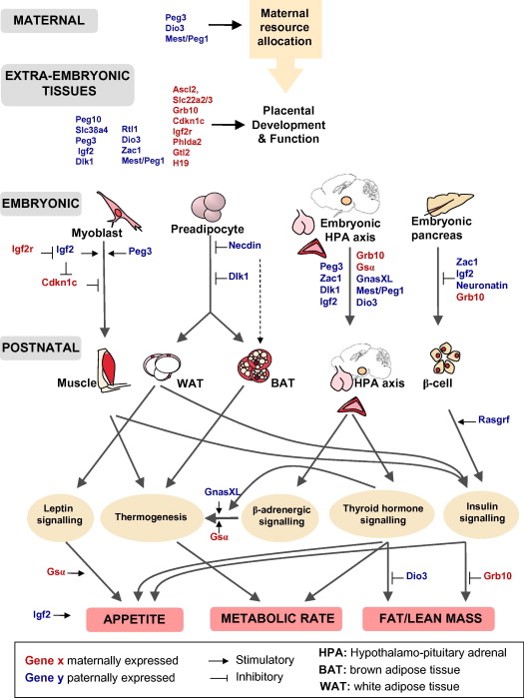
Figure 1. Imprinted genes are critical for the control of energy balance. The correct imprinted gene expression dosage is vital for energy homeostasis during both prenatal and postnatal life. Imprinted genes play critical roles in the control of foetal nutrient supply through effects on maternal metabolism and energy partitioning, placental development and function. There is also evidence that imprinted genes act coordinately in the foetus to regulate growth, thus altering foetal demand for maternal resources. Imprinted genes play key roles in the development of metabolic organs and modulate key adult metabolic pathways. From Radford EJ, Ferrón SR, Ferguson-Smith AC (2011) Genomic imprinting as an adaptative model of developmental plasticity. FEBS Lett. 585:2059-66 |
Our work (Al Adhami et al., Genome Res., 2015) supports an alternative mechanism in which imprinted genes cooperatively control a single biological process, which impacts on the function of various organs involved in the control of organismal growth. Using data from meta-analyses of microarray data, we showed that imprinted genes are frequently coexpressed (Figure 2).
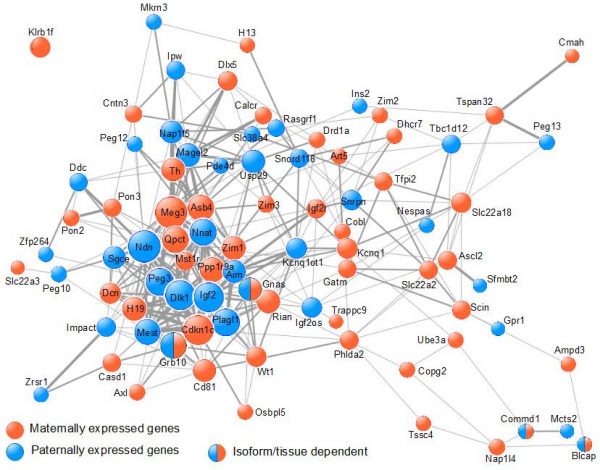
Figure 2. Imprinted genes are frequently co-expressed. The COXPRESdb meta-analysis of micro-array data was searched for co-expression among murine IGs. The resulting co-expression links are represented using Cytoscape. Node size is proportional to node degree. Edge width represents the mutual rank between 2 given nodes. |
To demonstrate that the observed imprinted gene co-expression is significant, we compared the topology of this network to the ones obtained with sets of genes known to be functionnally related, i.e. genes that belongs to the same ‘Biological Process’ as defined by Gene Ontology, or to sets of randomly selected genes (Figure 3)
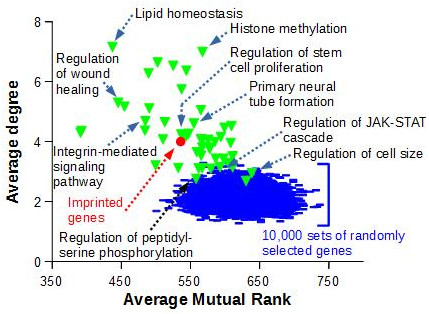
Figure 3. The average degree and average mutual rank were computed for the network of 85 imprinted genes displayed in Figure 2 (red circle). Random networks (blue lines) were generated by randomly drawing 10,000 sets of 85 GeneIDs with data in COXPRESdb, and retrieving co-expression links as for the 85 murine imprinted genes. A similar work was performed with 50 sets of 85 genes comprised in GO Biological Processes (green triangles) and with data in COXPRESdb |
We also showed that imprinted genes are coordinately regulated at the transition from proliferation to quiescence and differentiation during fibroblast cell cycle withdrawal, adipogenesis in vitro (Figure 4), and muscle regeneration in vivo.
|
Imprinted gene regulation is not linked to alteration of DNA methylation or to perturbation of mono-allelic, parent-of-origin-dependent expression. Overexpression and knockdown of imprinted gene expression alters the sensitivity of preadipocytes to contact inhibition and adipogenic differentiation (Figure 5).
|
In silico and in cellulo experiments showed that the imprinted gene network includes bi-allelically expressed, non-imprinted genes, which control the extracellular matrix composition, cell adhesion, cell junction, and extracellular matrix-activated and growth factor-activated signaling. These observations show that imprinted genes share a common biological process that may account for their seemingly diverse roles in embryonic development, obesity, diabetes, muscle physiology, and neoplasm. Future studies will further characterize the links between imprinted genes and extracellular matrix genes.
 Figure 4. Heat-map of imprinted gene expression levels during 3T3-L1 adipogenic differentiation. 3T3-L1 preadipocytes were grown exponentially (P, proliferation) until they reached confluence (Q, quiescence) following contact inhibition. Forty-eight hours later, they were induced to differentiate following addition of IDX (Insulin, Dexamethasone, IBMX), resumed proliferation during the clonal expansion phase (CE, clonal expansion), and eventually exited the cell cycle and differentiated (D, differentiation). Expression levels of the indicated imprinted genes were monitored by real-time PCR and expressed as the % of the maximal expression levels for that gene. Cebpd, Pparg, Lpl, Cebpa, Plin1, Adipoq, Slc2a4, and Lep are markers of early and late adipogenic differentiation.
Figure 4. Heat-map of imprinted gene expression levels during 3T3-L1 adipogenic differentiation. 3T3-L1 preadipocytes were grown exponentially (P, proliferation) until they reached confluence (Q, quiescence) following contact inhibition. Forty-eight hours later, they were induced to differentiate following addition of IDX (Insulin, Dexamethasone, IBMX), resumed proliferation during the clonal expansion phase (CE, clonal expansion), and eventually exited the cell cycle and differentiated (D, differentiation). Expression levels of the indicated imprinted genes were monitored by real-time PCR and expressed as the % of the maximal expression levels for that gene. Cebpd, Pparg, Lpl, Cebpa, Plin1, Adipoq, Slc2a4, and Lep are markers of early and late adipogenic differentiation. 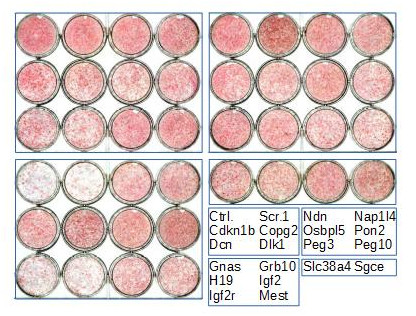 Figure 5. Effect of imprinted gene down-regulation on adipogenic differentiation of 3T3-L1 preadipocytes. Representative Oil Red O (ORO, a lipid stain) staining of 3T3-L1 cells that were left untransfected (Ctrl.) or transfected with scramble siRNA (Scr.1) or with siRNAs targeting the indicated genes, plated in duplicate at confluence, incubated with IDX 3 days post-transfection, and fixed 12 days later.
Figure 5. Effect of imprinted gene down-regulation on adipogenic differentiation of 3T3-L1 preadipocytes. Representative Oil Red O (ORO, a lipid stain) staining of 3T3-L1 cells that were left untransfected (Ctrl.) or transfected with scramble siRNA (Scr.1) or with siRNAs targeting the indicated genes, plated in duplicate at confluence, incubated with IDX 3 days post-transfection, and fixed 12 days later.